Cyanobacterial dynamics are a popular research topic for limnologists focused on understanding the patterns and behaviors of harmful algal blooms. These blooms can have long-lasting impacts on aquatic environments, such as general system health, altered food webs, and regional economic losses. These harms are worsened by anthropogenic factors like climate change and eutrophication.
With these compounding variables expected to continue to worsen, understanding long-term cyanobacterial dynamics is vital in considering resource management and future reliance on aquatic systems. A 2023 study published in Freshwater Biology sought to compare historical data on cyanobacterial dynamics and determine whether climate factors and the experimental stressors of eutrophication and acidification were associated with changes in cyanobacteria.

Image of Lake 239 at IISD Experimental Lakes Area from above (Credit: Sbath iisd via Wikimedia Commons CC BY-SA 4.0)
Methods
In order to examine the cyanobacterial dynamics and distinguish natural from anthropogenic influences, researchers focused on four lakes in Northwestern Ontario, Canada. Two of the lakes were experimentally manipulated (L227 and L223), and the other two were reference lakes (L224 and 442). Lakes in both groups had long-term phytoplankton monitoring records and known histories of various anthropogenic stressors. Data was extracted by analyzing sediment DNA (sedDNA) at the International Institute for Sustainable Development – Experimental Lakes Area.
Sediment DNA (sedDNA) was collected using core chronology, wherein a freeze corer was used to collect sediment cores in March 2018 near the maximum depths of the four lakes surveyed. The use of sedDNA in the study was validated by comparing results to other known methods of examining cyanobacteria dynamics, Droplet Digital Polymerase Chain Reaction (ddPCR) and High-Throughput Sequencing.
Researchers quantified three cyanobacterial genes using ddPCR. These included:
- The cyanobacterial 16S rRNA gene (CYA): A highly conserved small subunit rRNA gene found in all cyanobacteria. This gene was used as a proxy for total cyanobacterial abundance.
- The Microcystis 16S rRNA gene (MICR): A gene fragment specific to Microcystis—a common genus in freshwater environments.
- Microcystin E (mcyE): “A gene that is part of the operon involved in the production of the micro-cystin toxin.”1
SedDNA was assessed alongside coinciding environmental factors like temperature, total annual precipitation, heating degree days, cooling degree days, TP and TN. Sediment core data tracked cyanobacteria community demographics and water data from similar timeframes.
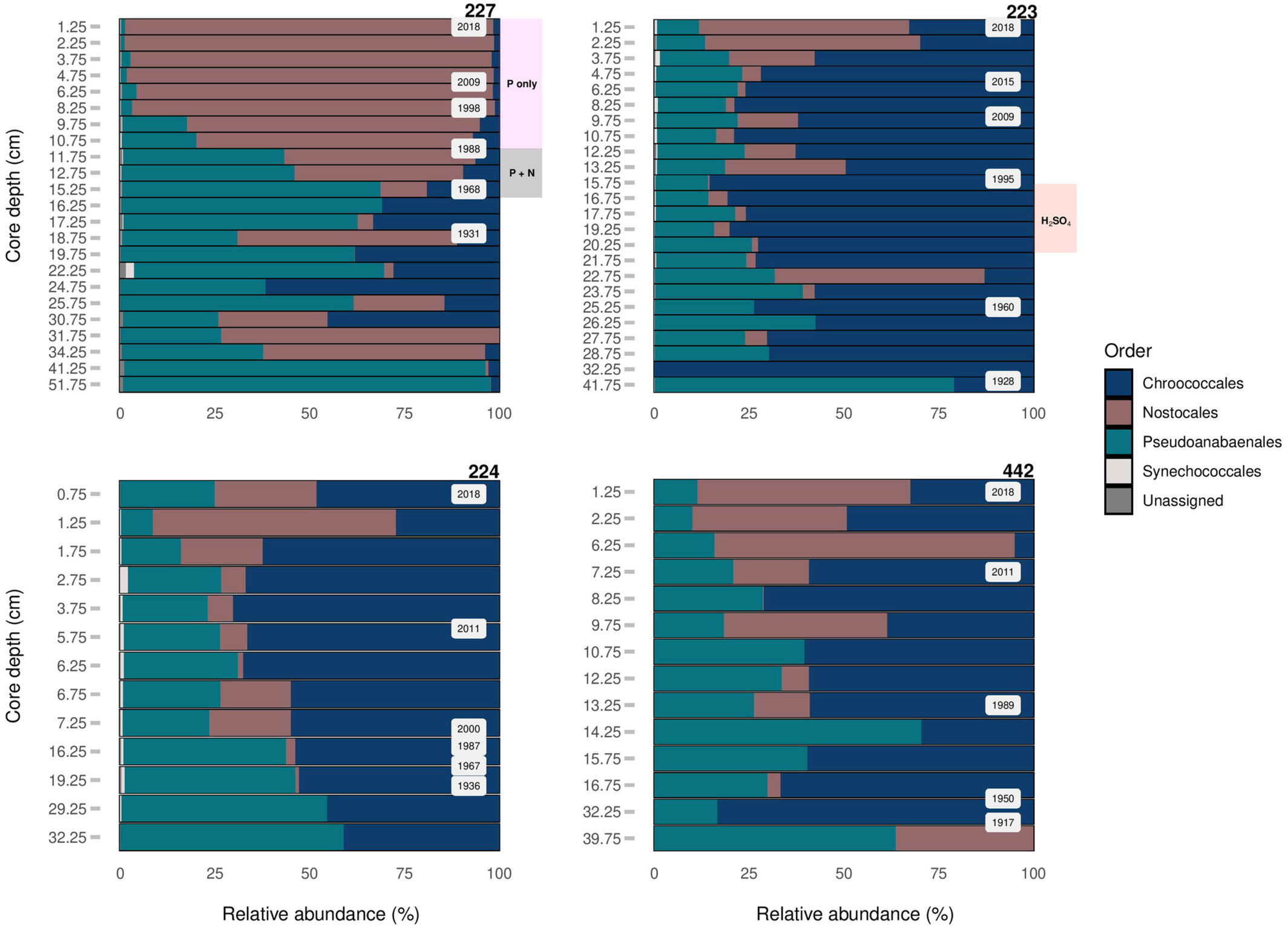
The reconstructed cyanobacterial community composition in lake sediment cores of the Experimental Lakes Area, Canada, from manipulated lakes 227 (top left) and 223 (top right), along with reference lakes 224 (bottom left) and 442 (bottom right), as determined through high‐throughput sequencing of the cyanobacterial 16S rRNA gene and using the Greengenes reference gene database. The relative abundance of major taxa is presented at the level of order. The grey shading in the Lake 227 plot (top left), labelled P + N, corresponds to the period of phosphorus and nitrogen loading in the lake between 1969 and 1989. The purple shading in the same plot, labelled P only, corresponds to the period of P loading only from 1990 to 2018. The pink shading in the Lake 223 plot (top right), labelled H2SO4, corresponds to the period of sulfuric acid loading in the lake between 1976 and 1993 (Credit: Mejbel et al., 2022)
Results
Samples revealed that all target genes showed an increase in abundance over time. Phytoplankton counts coincided with various water quality factors like total nitrogen or phosphorous counts. L227 underwent experimental eutrophication, and the change in water quality led to an increase in cyanobacteria production. Likewise, L223 underwent experimental acidification, which had similar effects, though to a lesser extent.
Comparing sedDNA and historical water column estimates revealed general congruence between the two methods in their ability to estimate community composition and the inferred cyanobacterial community structure. Analysis of coinciding environmental data showed that nutrients and climate variables correlated with long-term cyanobacterial dynamics. The influence of environmental factors like temperature was quantified by the reference lakes, which still experienced an increase in cyanobacteria abundance. SedDNA was less accurate in its estimates of specific phytoplankton species present, making other methods preferred.
In the reference lakes, heating degree days were a good indicator of shifts and variations in cyanobacterial dynamics. Such findings suggest that global climate change will lead to long-term changes in cyanobacterial community structures, potentially increasing toxigenic strains.
Sources:
- Mejbel, H. S., Irwin, C. L., Dodsworth, W., Higgins, S. N., Paterson, M. J., & Pick, F. R. (2023). Long-Term Cyanobacterial Dynamics from Lake Sediment DNA In Relation To Experimental Eutrophication, Acidification and Climate Change. Freshwater Biology, 00, 1– 19. https://doi.org/10.1111/fwb.14074







