Water quality monitoring can be challenging for regions with limited access to resources, time and trained professionals. In the past, this issue has largely been ignored, leading to infrequent or non-existent sampling. In areas like Lake Tanganyika, where sanitation is limited to pit latrines, pour-flush latrines, flushing toilets with septic tanks and limited incidences of open defecation, sampling water for coliforms and other bacteria can make a significant difference in coliform concentrations and overall water quality.1
Coliform bacteria are non-pathogenic bacteria that are present in the feces of warm-blooded animals. When found in large concentrations, their presence is associated with various waterborne diseases, such as gastrointestinal illness, typhoid fever, ear infections, diarrhea and dysentery.2 Coliforms may also be seen as a probable threat or indicator of deterioration of microbiological water quality.3

Lake Tanganyika. (Credit: Dave Proffer via Flickr CC BY 2.0)
Understanding the potential threats that high coliform concentrations pose, inconsistent or non-existent monitoring can leave communities unknowingly exposed to various diseases. In order to bridge this gap in environmental monitoring, citizen science initiatives can encourage public engagement and create a more comprehensive view of water quality.
While some have questioned the accuracy and reliability of citizen science programs, these projects have the potential to build quality data at a fraction of the cost. A 2022 study published in PLOS ONE assessed the use of citizen science in monitoring the dynamics of coliform concentrations in the Lake Tanganyika region as a complementary method to statutory measurements conducted by scientists.
Methods
Individuals from five coastal communities were chosen and trained to monitor total coliforms, fecal coliforms and turbidity for one year on a monthly basis. These measurements were conducted in tandem with professional scientists. The two groups used the same methods to sample and assess coliform concentrations so that the results of each group’s analysis could be compared.
The five coastal communities chosen represented various contaminant output levels in order to test the reliability of citizen science programs accurately.
- Ilagala has a high population size and receives effluents from the Malagarasi River. The sediment load from the Malagarasi is elevated during the rainy season.
- Kibirizi has an important fishery industry, with the largest fish market being included in one of the community’s sampling sites. It is also in a closed bay, leading to a higher retention in runoff. The streams that feed into this river carry effluent from areas with small population centers and agricultural activities.
- Gombe is a small community located near a national park. Most economic activities in the park are prohibited, so the area is well-vegetated and conserved. The healthy natural landscape reduces possible influxes of livestock and human waste.
- Karago is a small rural area in a semi-closed bay. The community has a low population and animal density. Evidence suggests that large quantities of wastewater do not flow directly into the area during heavy rain events. Similar to Gombe, Karago had low economic and agricultural activities.
- Ujiji “presents mixed conditions with high coliforms and turbidity, but a low influence from precipitation.”1 It has a population similar to that of Ilagaala and is located near the discharge of the Luiche River and several agricultural villages.

Beach at day Bangwe Ward, Kigoma-Ujiji District, Kigoma Region, Tanzania. (Credit: Halidtz via Wikimedia Commons CC BY-SA 4.0)
Results
Results of the coliform sampling revealed there was a strong seasonal dynamic, with higher concentrations in the rainy seasons—a similar dynamic was observed for fecal coliforms. Seasonal dynamics varied from sit-to-site, with a clear difference in concentration between sampling sites. Variables that appeared to influence coliforms and turbidity were sites with higher economic activity, shorter distance from households, higher farming intensity, river proximity and urban dominance.1
The highest concentrations occurred in Ilagala and the lowest in Gombe and Karago. Turbidity concentrations also varied significantly between sites. Similar to coliform concentration, turbidity was the highest at Ilagala and lowest at Gombe.
Comparing the two sampling groups (citizen scientists and professional scientists) revealed a very strong correlation in results between the two groups with slight variation. For total coliforms, results were similar, with a minor overestimation of citizen scientists’ values over professional scientists’ values. Similarly, the fecal coliform results had a high correlation between the two groups. Again, there was a slight overestimation of the citizen scientists’ values.
While there were some inaccuracies, the correlation between the two groups is strong enough that the article suggests citizen scientist initiatives as a realistic solution to filling data gaps. When well-trained and properly equipped, citizen scientists can be a valuable asset to the community in discovering unsafe conditions and to researchers hoping to gain more knowledge of these lakes.
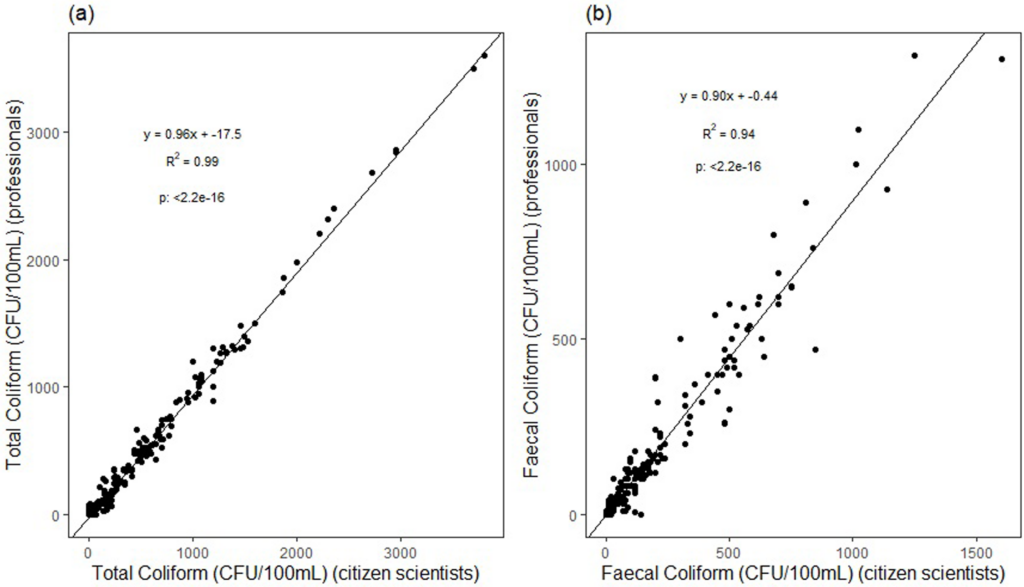
Pearson correlation of total coliform and fecal coliform concentration (CFU/100 mL) observed by professional and citizen scientists (a) Total coliform (CFU/100 mL) (b) fecal coliform (CFU /100m/L) (n = 180). (Credit: Moshi et al., 2022)
Sources
- Moshi HA, Shilla DA, Kimirei IA, O’ Reilly C, Clymans W, et al. (2022) Community monitoring of coliform pollution in Lake Tanganyika. PLOS ONE 17(1): e0262881. https://doi.org/10.1371/journal.pone.0262881
- Gruber JS, Ercumen A, Colford JM Jr. Coliform bacteria as indicators of diarrheal risk in household drinking water: systematic review and meta-analysis. PloS One. 2014;9(9):e107429. pmid:25250662
- Ndlovu T, Le Roux M, Khan W, Khan S. Co-detection of virulent Escherichia coli genes in surface water sources. PloS One. 2015;10(2):e0116808. pmid:25659126